
Introduction to R
What? And why?
R is a language developed in an open-source environment. It is an interpreted language that does not need to be compiled by a compiler, and allows rapid programming with its interactive command-line interface (similar to the Python one) and scripting.
Due to its straightforwardness, it is widely used in data mining and data analysis. As one of the most commonly used numerical computing programming languages, it is worth having a brief introduction to it.
RStudio
We hardly ever just write code from the command line, and even less if the language we are using is mainly to get visual analysis of the data. Therefore, the most common way to use R is by Rstudio, a Graphical User Interface software to develop R code.

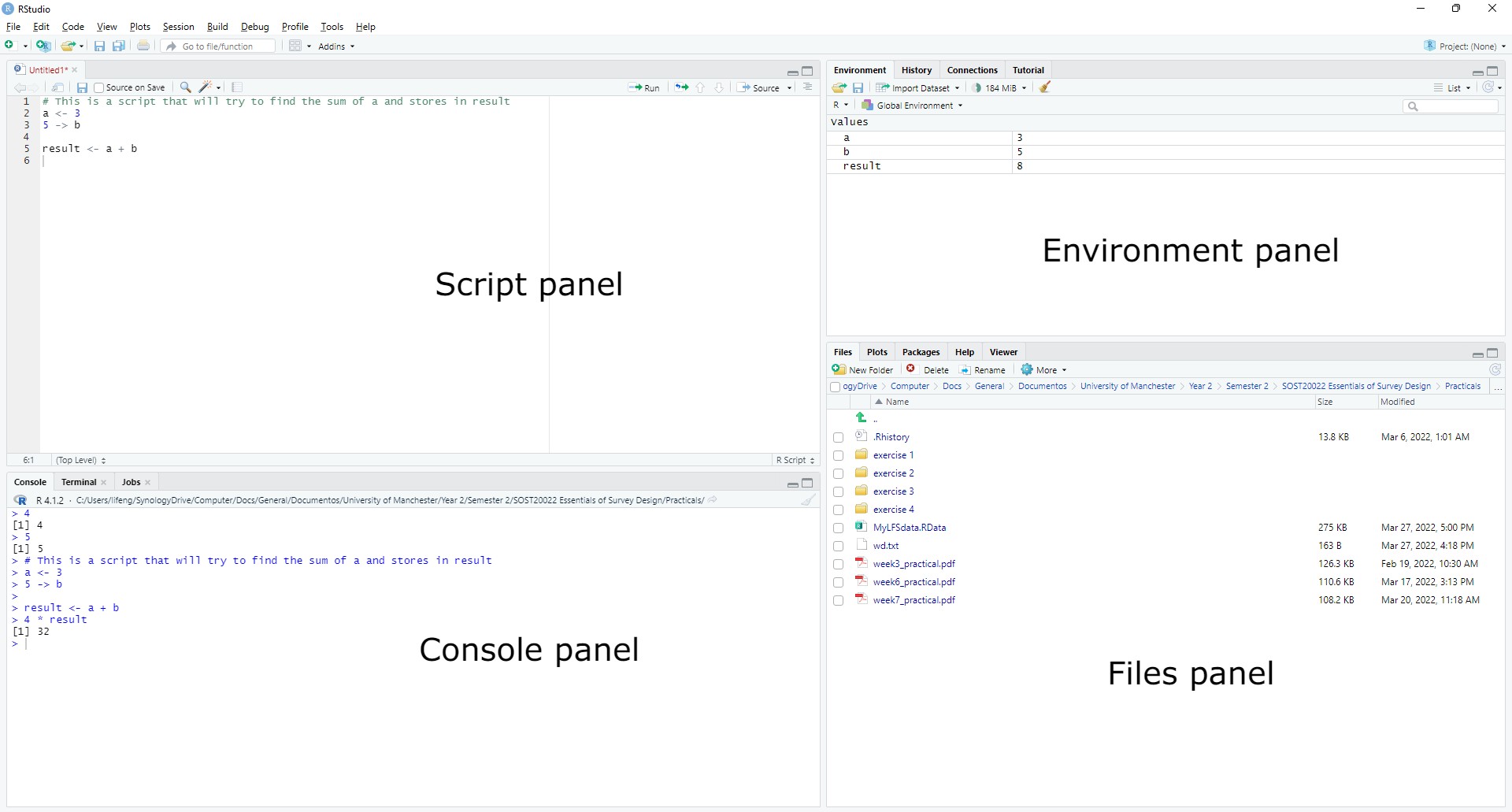
Once downloaded and inside, 4 panels (3 if the script panel is not opened) will show up. From left top to right bottom, they are:
- Script panel: where you can write blocks of code to be executed. A very helpful functionality that I find, is that it allows you to select the lines of codes that you want to run if not all, by selecting them and pressing run.
- Console panel: where the interactive terminal is. You can either enter code here to be executed or use a script. The outputs e.g. print outputs will be shown here.
- Environment panel: where you can see all the variables stored currently.
- Another useful tab in this panel is history, where you can check all previously executed commands.
- Files panel: where you can see the files in a explorer format.
- Plots tab allows you to visualise output of any R figures.
- Packages to explore any installed R packages.
- Help outputs the relevant information when help(“some_command”) is called
R packages
R as a community comes with the huge support of packages. Install a package with
1 | install.packages("package_name") |
Incorporate packages into your library with
1 | library("package_name") |
These actions can also be done in the RStudio GUI.
R scripts
Very often, we need to use a piece of code on multiple occasions, that’s why it is useful to store them in a file. These files are called R scripts.
1 | # This is a script that calculates the root mean square of two numbers |
Loading data
R‘s default reading function is read.table, and is a bit limited in the way that it can only read human-readable data files (e.g. tab-separated, comma separated). The result is a data.frame object.
1 | data <- read.table(filename, header=TRUE, sep="\t") |
Data structures
Vector
Vector is a group of values ordered as an array in which all elements should be of the same type.
There are several ways of instantiating a vector:
1 | v <- c(1, 2, 3, 4, 5) |
To extract from a vector
1 | v[1] # first element |
List
List is a vector that allows multiple types of values.
List follows the same creation and extraction rules as a vector, only that each element is also considered as a list. Hence:
1 | l <- list(1, "a", TRUE, c(-1, -2, -3)) |
Matrix
Matrix is a vector of vectors; a 2D vector, and has a rectangular shape.
Create a matrix
1 | m <- matrix(c(1, 2, 3, 4, 5, 6), nrow=2) |
Extract an element or a vector from a matrix
1 | m[1, 2] # first row, second column; 3 |
Data frame
Data frame is the most commonly used data structure in R. It is a matrix in which each column is considered an attribute of the data, and each row is considered an observation. As attributes, columns always have their name. If not declared explicitly, they will be called X1, X2, X3, etc.
For instance, in a survey, each row is a person, and each column is a variable from a question (e.g. age, sex, income).
The read.table automatically loads data into a data frame. Other way of creating one is from a matrix.
1 | df <- data.frame(m) |
The extraction follows the same rules as for matrices. One difference is in the extraction of columns with df[, 2] and df[2]. Notice as we have a name for each of the columns, we can extract the column with the name.
1 | df[, 2] # if with 2 coordinate system, the result is flattened |
You may have guessed, names in column is fairly important, here is how to change them:
1 | names(df) <- c("new_name_1", "new_name_2", "new_name_3") # change all names |
Boolean and masks
Data structures can interact with boolean values, providing filter functions (mask).
1 | # runif(n) creates n random numbers between 0 and 1 |
Other common syntax
R has a very straightforward syntax, some of its regular syntax example are the following.
Loops
1 | for (i in 1:10) { |
Conditionals
1 | if (i > 5) { |
Functions
1 | plus_one(x) { |
Visualisation packages
A good place to start is the ggplot2 package.
R Markdown
R Markdown is a simple syntax for writing R code in a Markdown-like format.
Conclusion
R is a very powerful language, and it is very easy to learn. This has been a quick introductory post about its general syntax and some of the most common features. The next R tutorial may be a real example of how to use it to extract wanted data and perform statistical analysis on it.
- Title: Introduction to R
- Author: Gnefil Voltexy
- Created at : 2022-04-13 20:11:09
- Updated at : 2025-10-02 09:51:20
- Link: https://blog.gnefil.com/2022-04-13/Introduction-to-R/
- License: This work is licensed under CC BY-SA 4.0.
